Note
Click here to download the full example code
Isolated cluster and bad init¶
The iterative process of Active Learning may induce corner cases that are proper to Active Learning. In this case, we look at what happens if some data has an isolated cluster where no point is selected at init.
This cluster may be totally overlooked by some samplers that do not explore the dataset well enough through diversity or representativity sampling.
from copy import deepcopy
import numpy as np
from sklearn.datasets import make_blobs
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
from matplotlib import pyplot as plt
from cardinal.utils import ActiveLearningSplitter
from cardinal.uncertainty import ConfidenceSampler
from cardinal.random import RandomSampler
from cardinal.clustering import KMeansSampler
from cardinal.plotting import smooth_lines
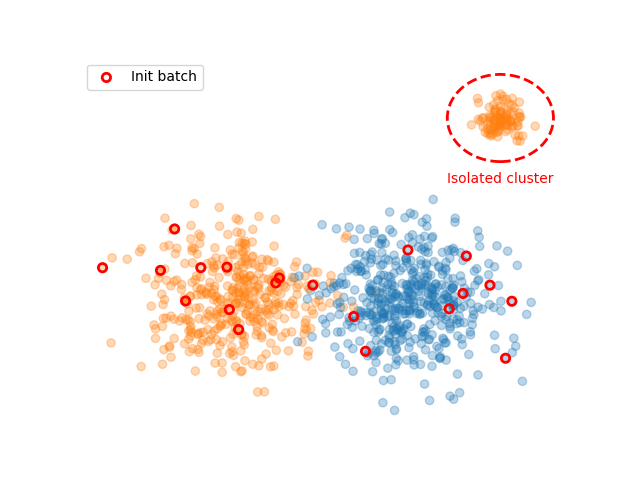
We simulate two classes. Class 0 is simply a large blob. Class 1 is composed of a large blob and an isolated cluster located aside. In order to generate this cluster, the data for this class is seperated into two blobs.
X, blob = make_blobs([500, 400, 100], centers=[(2, 0), (-2, 0), (4, 5)], cluster_std=[1.0, 1.0, 0.3], random_state=0)
batch_size = 10
clf = LogisticRegression()
We now create the active learning experiment. We use cardinal’s splitter to handles indices. For the initialisation, we only sample data from the first two blobs. This simulates an unlucky initialization where no data from the isolated cluster is selected.
init_spl = ActiveLearningSplitter.train_test_split(X.shape[0], test_size=0.2, stratify=blob, random_state=0)
init_idx = np.hstack([
np.where(blob[init_spl.train] == 0)[0][:batch_size],
np.where(blob[init_spl.train] == 1)[0][:batch_size]
])
init_spl.initialize_with_indices(init_idx)
left_out_y = (blob[init_spl.test] == 2)
y = blob.copy()
y[blob == 2] = 1
plt.scatter(X[:, 0], X[:, 1], c=['C{}'.format(i) for i in y], alpha=.3)
plt.scatter(X[init_spl.selected, 0], X[init_spl.selected, 1], facecolors='none', edgecolors='r', linewidth=2, label='Init batch')
plt.gca().add_patch(plt.Circle((4, 5), 1.2, color='r', fill=False, linestyle='dashed', linewidth=2))
plt.text(4, 3.2, 'Isolated cluster', ha='center', c='r')
plt.legend()
plt.axis('off')
plt.show()
This function runs the experiment. It is a class active learning setting.
def evaluate(name, sampler, g_ax, ic_ax):
spl = deepcopy(init_spl)
g_acc = []
ic_acc = []
for _ in range(10):
clf.fit(X[spl.selected], y[spl.selected])
sampler.fit(X[spl.selected], y[spl.selected])
spl.add_batch(sampler.select_samples(X[spl.non_selected]))
g_acc.append(accuracy_score(y[spl.test], clf.predict(X[spl.test])))
ic_acc.append(accuracy_score(y[spl.test][left_out_y], clf.predict(X[spl.test][left_out_y])))
g_ax.plot(np.arange(10), g_acc, label=name)
ic_ax.plot(np.arange(10), ic_acc, label=name)
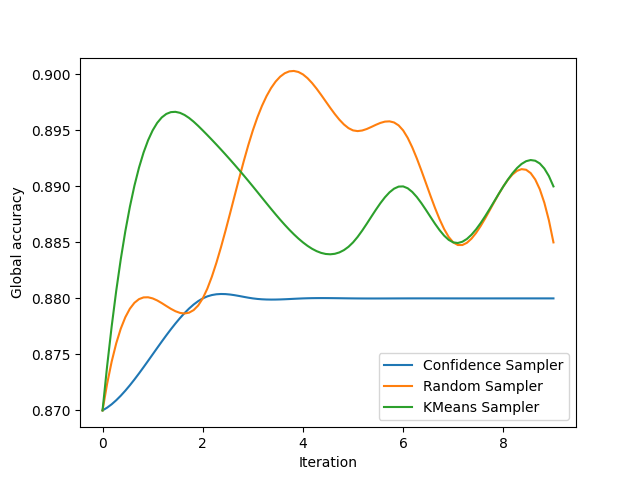
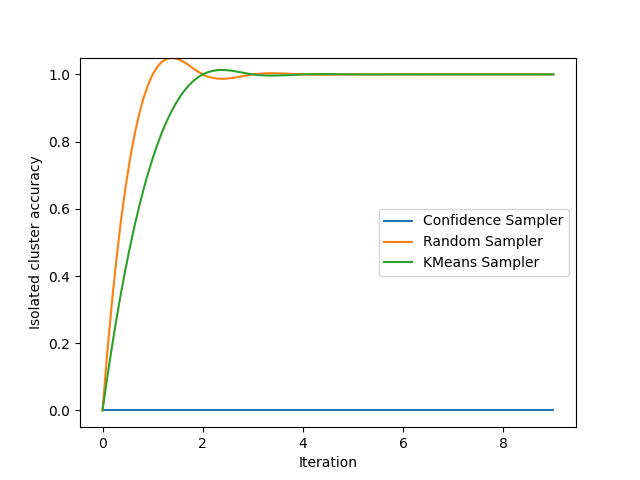
We now display the results for 3 very common samplers. You may observe that the confidence sampling completely ignores the isolated cluster since it is designed to focus on the existing decision boundary. Random sampling has a 15% chance of picking a sample in this cluster during the experiment. By design, KMeans sampling will always select samples in the isolated cluster!
plt.figure()
global_ax = plt.gca()
plt.ylabel('Global accuracy')
plt.xlabel('Iteration')
plt.figure()
isolated_cluster_ax = plt.gca()
plt.ylabel('Isolated cluster accuracy')
plt.xlabel('Iteration')
evaluate('Confidence Sampler', ConfidenceSampler(clf, batch_size=batch_size, assume_fitted=True), global_ax, isolated_cluster_ax)
evaluate('Random Sampler', RandomSampler(batch_size=batch_size, random_state=0), global_ax, isolated_cluster_ax)
evaluate('KMeans Sampler', KMeansSampler(batch_size=batch_size), global_ax, isolated_cluster_ax)
global_ax.legend()
smooth_lines(axis=global_ax, k=3)
isolated_cluster_ax.legend()
smooth_lines(axis=isolated_cluster_ax, k=3)
plt.show()
Total running time of the script: ( 0 minutes 4.263 seconds)
Estimated memory usage: 9 MB